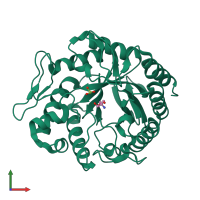
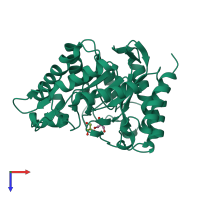
Function and Biology Details
Reaction catalysed:
Random hydrolysis of (1->4)-beta-D-mannosidic linkages in mannans, galactomannans and glucomannans
Biochemical function:
Biological process:
Cellular component:
Structure analysis Details
Assembly composition:
monomeric (preferred)
Assembly name:
Mannan endo-1,4-beta-mannosidase A (preferred)
PDBe Complex ID:
PDB-CPX-109480 (preferred)
Entry contents:
1 distinct polypeptide molecule
Macromolecule: